Quick Summary
 Highly efficient statistician always looking into creating a meaningful impact. Over my experience in mostly research environments across Europe and Australia, I am able to take innovative initiative to lead projects to successful completion within timeline and budget with deliverables that are understandable by non-specialists.
Highly efficient statistician always looking into creating a meaningful impact. Over my experience in mostly research environments across Europe and Australia, I am able to take innovative initiative to lead projects to successful completion within timeline and budget with deliverables that are understandable by non-specialists. Based in Brisbane-Australia since some day of Automn 2014 (harsh season with its 20-25 degrees and around 8 rainy days a month), I'm always up for a new challenge locally or internationally.
Skills - Keywords
Non-exhaustive list of my skills:Statistics Statistical Data Analysis Applied Mathematics Mathematical Modeling Project Management Big Data Machine Learning Data Science Forecasting Predictive Analytics Predictive Modeling Data Visualization Data Mining Algorithms Algorithms Development R Git LaTeX Teaching
Greatest research outcomes
• Creation of the ’Rohart MSC test’ - identification of a set of 16 markers that improved the recognition of stem cells with potential for clinical use (human mesenchymal stromal cells MSC) via proactive development of a new innovative machine learning approach.The test is used internationally online, and you can have a direct look at what the graphical outputs looks like on a time course data where you can see the loss of the MSC function with our test, pretty cool uh?
The novel statistical approach is implemented in a R-package bootsPLS, available on CRAN; it imports the mixOmics package and perform subsampling to identify a stable signature.
 • mixOmics: an R package for `omics feature selection and multiple data integration. I have developed mixOmics from v6.00 to v6.8 (included) with the major developments of the MINT and DIABLO modules (added in v6.0.0), as well as a huge gain in computational time and memory usage from v6.3.0 onwards. For instance, one of the core functions (splsda) is now more than 100x faster when there is missing data (~15x faster with no missing data); memory usage is down 7-17x. I am still involved in mixOmics (at the time of writing) but this is not my main occupation at the moment.
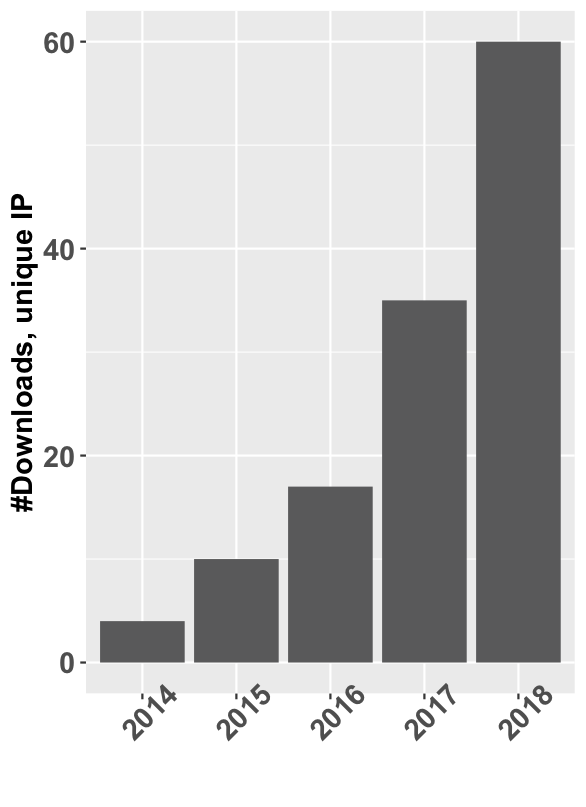
• mixOmics: an R package for `omics feature selection and multiple data integration. I have developed mixOmics from v6.00 to v6.8 (included) with the major developments of the MINT and DIABLO modules (added in v6.0.0), as well as a huge gain in computational time and memory usage from v6.3.0 onwards. For instance, one of the core functions (splsda) is now more than 100x faster when there is missing data (~15x faster with no missing data); memory usage is down 7-17x. I am still involved in mixOmics (at the time of writing) but this is not my main occupation at the moment.mixOmics is continually growing, from barely 4k downloads in 2014 to around 60K in 2018 (cf plot). It is designed to be used for data analysis and data integration, there is a website with a lot of information and tutorials, a mailing list for updates and a github repository for issues.
Themes of interest
In lay terms, my research interests lie in extracting signal from big noisy data. That is mainly what I have been doing since the start of my research life in 2009. Usually that involves:- high dimensional linear (mixed) models
- variable selection
- multivariate analysis
- data integration
last modified: